
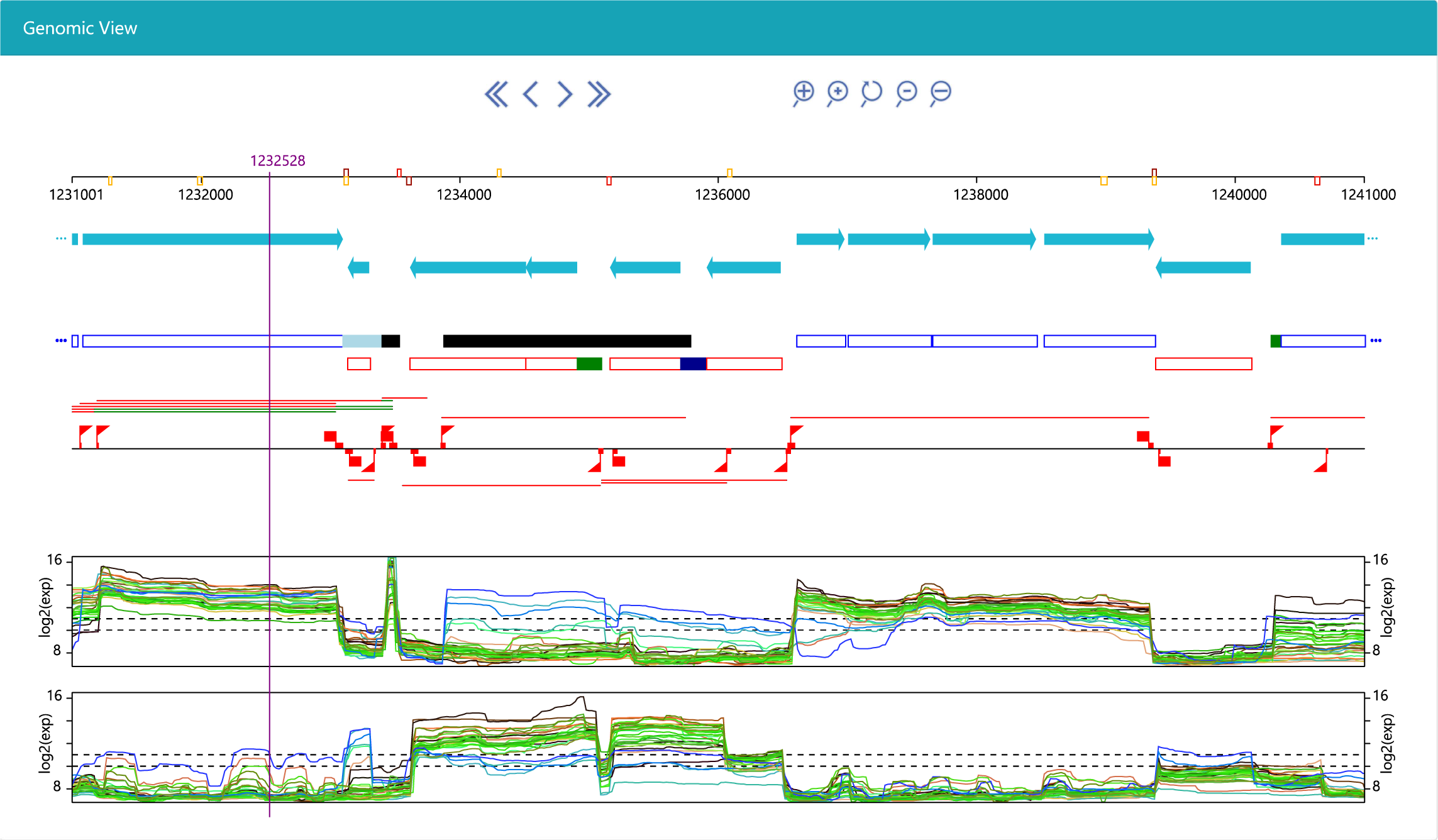
Genoscapist is a web-tool generating high-quality images for interactive visualization of hundreds of quantitative profiles along a reference genome together with various annotations.
Applications
- B. subtilis
- http://genoscapist.migale.inrae.fr/seb_bsubbiofilm
Presenting data from Direct comparison of spatial transcriptional heterogeneity across diverse Bacillus subtilis biofilm communities (Dergham et al., 2023) - http://genoscapist.migale.inrae.fr/smgeb
Presenting data from Greedy reduction of Bacillus subtilis genome yields emergent phenotypes of high resistance to DNA damaging agent and low evolvability (Dervyn et al., 2023) - http://genoscapist.migale.inrae.fr/seb_rho
Presenting data from Termination factor Rho mediates transcriptional reprogramming of Bacillus subtilis stationary phase (Bidnenko et al., 2023) - http://genoscapist.migale.inrae.fr/seb
Presenting transcriptome-based reannotation studie of Bacillus subtilis (Nicolas et al., 2012)
- http://genoscapist.migale.inrae.fr/seb_bsubbiofilm
- S. aureus
- http://genoscapist.migale.inrae.fr/aeb
Presenting transcriptome-based reannotation studie of Staphylococcus aureus (Mäder et al., 2016)
- http://genoscapist.migale.inrae.fr/aeb
- Virtual Machine running B. subtilis light dataset
Citation
Dérozier S, Nicolas P, Mäder U, Guérin C. Genoscapist: online exploration of quantitative profiles along genomes via interactively customized graphical representations [published online ahead of print, 2021 Feb 3]. Bioinformatics. 2021; btab079. doi:10.1093/bioinformatics/btab079.
Web-tool source code: https://doi.org/10.15454/OMDLOD.
Source code
The source code is available on a GitHub repository and placed under the GNU General Public License.
Users Mailing List
To keep users of the application informed of new features, we’ve set up a dedicated mailing list: genoscapist-users-request@groupes.renater.fr, which you can subscribe to by clicking here.